Vanadium »
PDB 7q0v-9g2m »
9dq2 »
Vanadium in PDB 9dq2: Crystal Structure of Hrmj From Streptomyces Sp. Cfmr 7 (Hrmj-Ssc) Complexed with Vanadyl(IV)-Oxo, Succinate and 6-Nitronorleucine
Protein crystallography data
The structure of Crystal Structure of Hrmj From Streptomyces Sp. Cfmr 7 (Hrmj-Ssc) Complexed with Vanadyl(IV)-Oxo, Succinate and 6-Nitronorleucine, PDB code: 9dq2
was solved by
Y.-C.Zheng,
W.-C.Chang,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 41.36 / 1.55 |
| Space group | P 21 21 21 |
| Cell size a, b, c (Å), α, β, γ (°) | 75.113, 78.836, 97.166, 90, 90, 90 |
| R / Rfree (%) | 19.6 / 21.1 |
Other elements in 9dq2:
The structure of Crystal Structure of Hrmj From Streptomyces Sp. Cfmr 7 (Hrmj-Ssc) Complexed with Vanadyl(IV)-Oxo, Succinate and 6-Nitronorleucine also contains other interesting chemical elements:
| Bromine | (Br) | 4 atoms |
Vanadium Binding Sites:
The binding sites of Vanadium atom in the Crystal Structure of Hrmj From Streptomyces Sp. Cfmr 7 (Hrmj-Ssc) Complexed with Vanadyl(IV)-Oxo, Succinate and 6-Nitronorleucine
(pdb code 9dq2). This binding sites where shown within
5.0 Angstroms radius around Vanadium atom.
In total 2 binding sites of Vanadium where determined in the Crystal Structure of Hrmj From Streptomyces Sp. Cfmr 7 (Hrmj-Ssc) Complexed with Vanadyl(IV)-Oxo, Succinate and 6-Nitronorleucine, PDB code: 9dq2:
Jump to Vanadium binding site number: 1; 2;
In total 2 binding sites of Vanadium where determined in the Crystal Structure of Hrmj From Streptomyces Sp. Cfmr 7 (Hrmj-Ssc) Complexed with Vanadyl(IV)-Oxo, Succinate and 6-Nitronorleucine, PDB code: 9dq2:
Jump to Vanadium binding site number: 1; 2;
Vanadium binding site 1 out of 2 in 9dq2
Go back to
Vanadium binding site 1 out
of 2 in the Crystal Structure of Hrmj From Streptomyces Sp. Cfmr 7 (Hrmj-Ssc) Complexed with Vanadyl(IV)-Oxo, Succinate and 6-Nitronorleucine
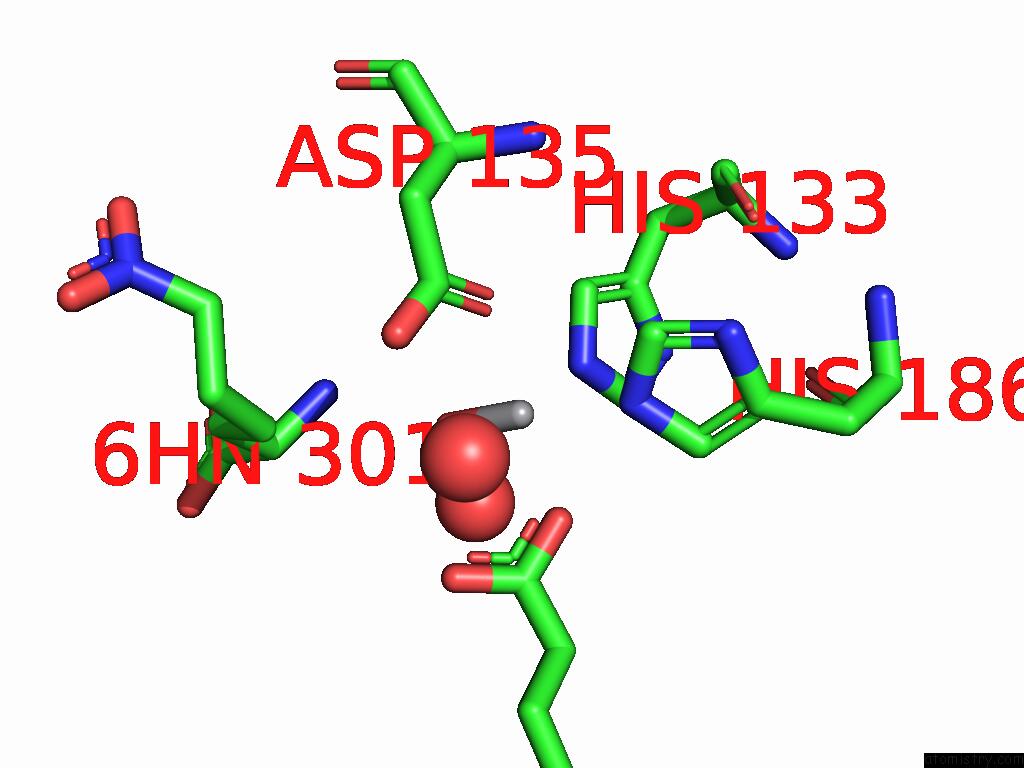
Mono view
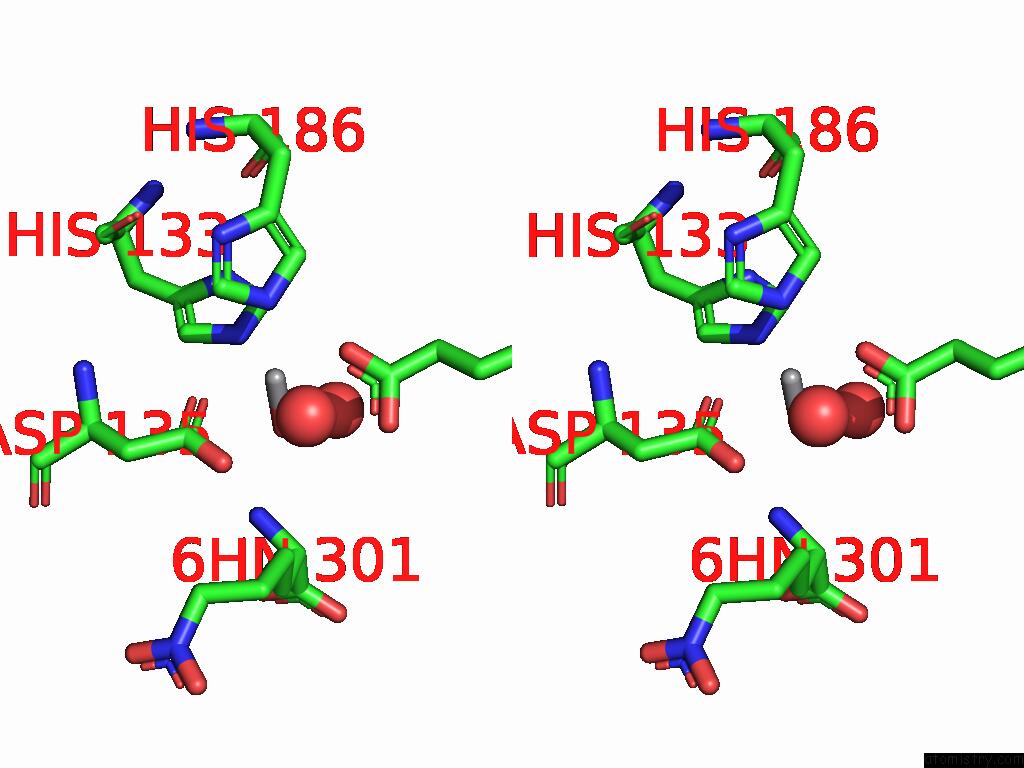
Stereo pair view

Mono view

Stereo pair view
A full contact list of Vanadium with other atoms in the V binding
site number 1 of Crystal Structure of Hrmj From Streptomyces Sp. Cfmr 7 (Hrmj-Ssc) Complexed with Vanadyl(IV)-Oxo, Succinate and 6-Nitronorleucine within 5.0Å range:
|
Vanadium binding site 2 out of 2 in 9dq2
Go back to
Vanadium binding site 2 out
of 2 in the Crystal Structure of Hrmj From Streptomyces Sp. Cfmr 7 (Hrmj-Ssc) Complexed with Vanadyl(IV)-Oxo, Succinate and 6-Nitronorleucine
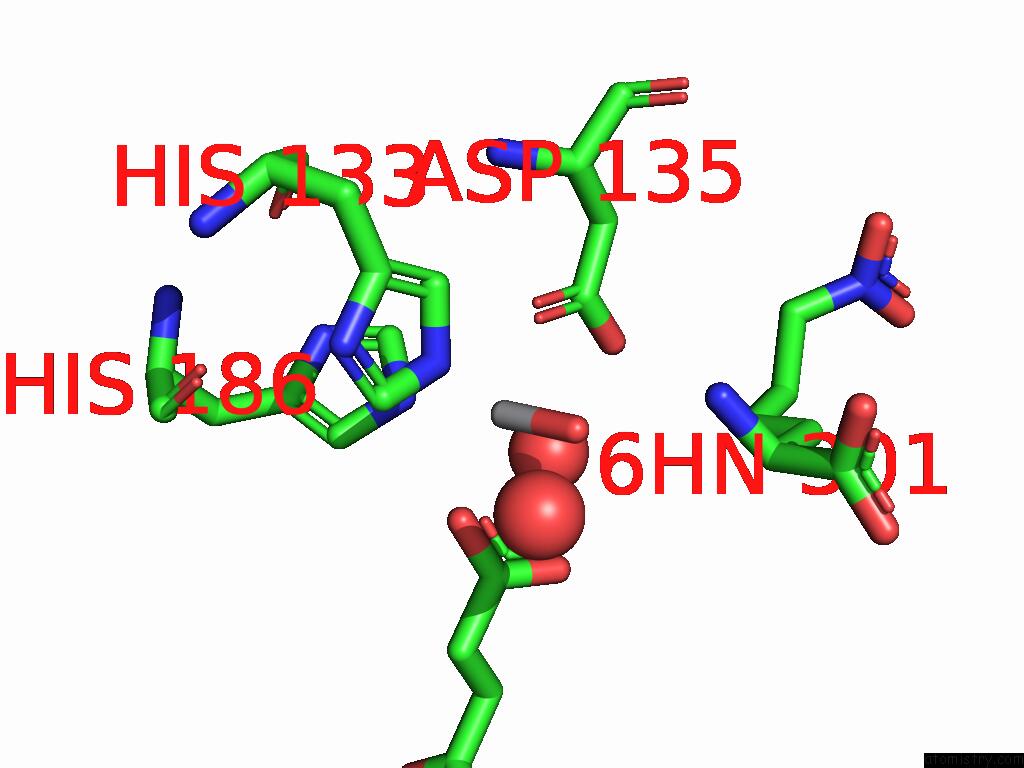
Mono view
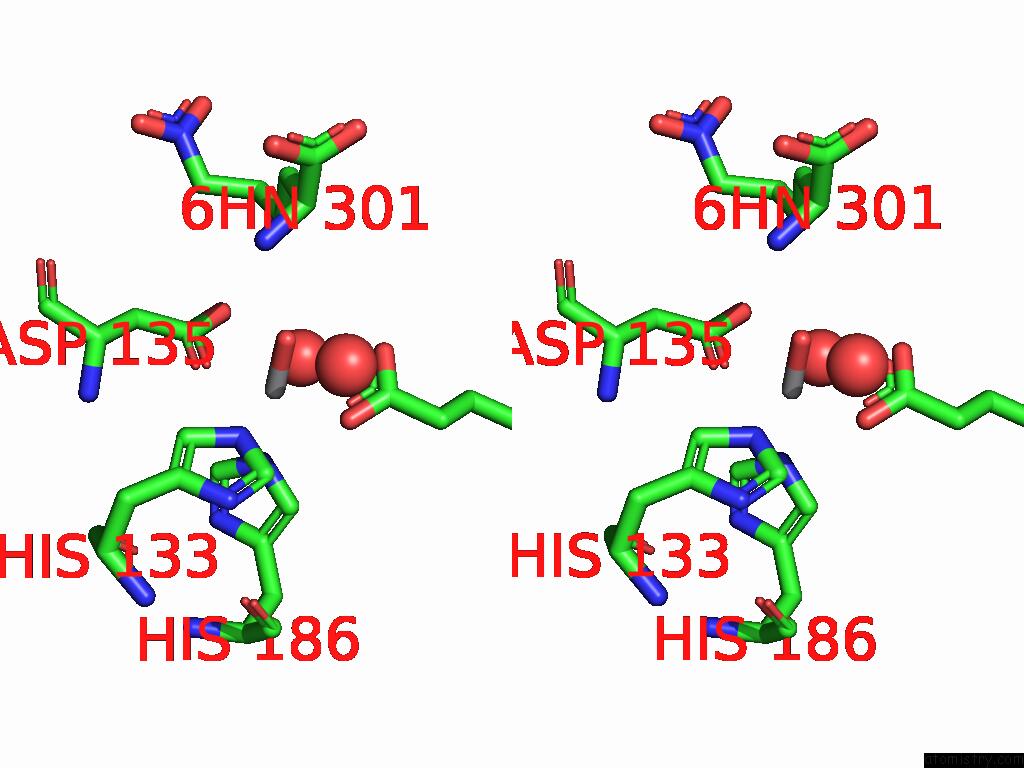
Stereo pair view

Mono view

Stereo pair view
A full contact list of Vanadium with other atoms in the V binding
site number 2 of Crystal Structure of Hrmj From Streptomyces Sp. Cfmr 7 (Hrmj-Ssc) Complexed with Vanadyl(IV)-Oxo, Succinate and 6-Nitronorleucine within 5.0Å range:
|
Reference:
Y.C.Zheng,
X.Li,
L.Cha,
J.C.Paris,
C.Michael,
R.Ushimaru,
Y.Ogasawara,
I.Abe,
Y.Guo,
W.C.Chang.
Comparison of A Nonheme Iron Cyclopropanase with A Homologous Hydroxylase Reveals Mechanistic Features Associated with Distinct Reaction Outcomes. J.Am.Chem.Soc. 2025.
ISSN: ESSN 1520-5126
PubMed: 39901767
DOI: 10.1021/JACS.4C17741
Page generated: Tue Feb 25 11:49:10 2025
ISSN: ESSN 1520-5126
PubMed: 39901767
DOI: 10.1021/JACS.4C17741
Last articles
I in 1PVHI in 1PGG
I in 1PNN
I in 1OXV
I in 1PGF
I in 1OXX
I in 1PGE
I in 1P59
I in 1OXS
I in 1ORW