Vanadium »
PDB 3omx-4zg4 »
4e0g »
Vanadium in PDB 4e0g: Protelomerase Tela/Dna Hairpin Product/Vanadate Complex
Protein crystallography data
The structure of Protelomerase Tela/Dna Hairpin Product/Vanadate Complex, PDB code: 4e0g
was solved by
K.Shi,
H.Aihara,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 40.12 / 2.20 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 117.880, 120.200, 58.040, 90.00, 111.85, 90.00 |
| R / Rfree (%) | 19.4 / 23.8 |
Vanadium Binding Sites:
The binding sites of Vanadium atom in the Protelomerase Tela/Dna Hairpin Product/Vanadate Complex
(pdb code 4e0g). This binding sites where shown within
5.0 Angstroms radius around Vanadium atom.
In total only one binding site of Vanadium was determined in the Protelomerase Tela/Dna Hairpin Product/Vanadate Complex, PDB code: 4e0g:
In total only one binding site of Vanadium was determined in the Protelomerase Tela/Dna Hairpin Product/Vanadate Complex, PDB code: 4e0g:
Vanadium binding site 1 out of 1 in 4e0g
Go back to
Vanadium binding site 1 out
of 1 in the Protelomerase Tela/Dna Hairpin Product/Vanadate Complex
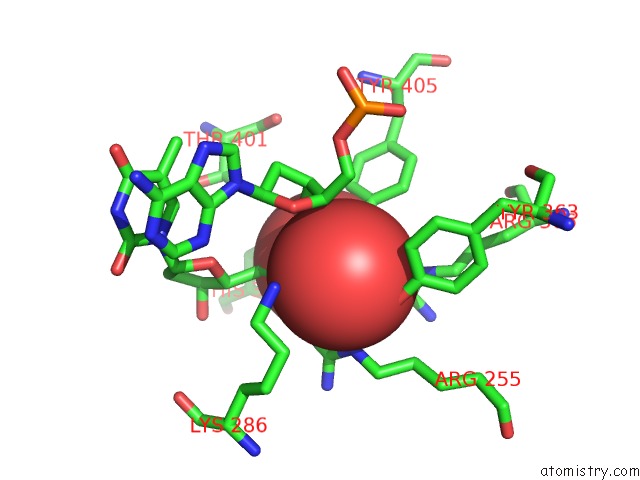
Mono view
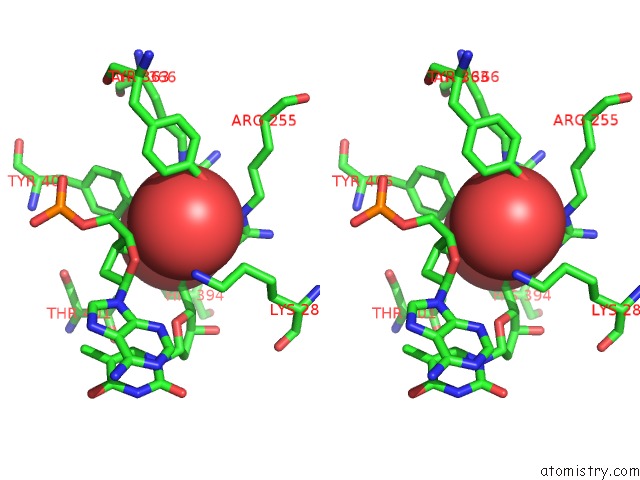
Stereo pair view

Mono view

Stereo pair view
A full contact list of Vanadium with other atoms in the V binding
site number 1 of Protelomerase Tela/Dna Hairpin Product/Vanadate Complex within 5.0Å range:
|
Reference:
K.Shi,
W.M.Huang,
H.Aihara.
An Enzyme-Catalyzed Multistep Dna Refolding Mechanism in Hairpin Telomere Formation. Plos Biol. V. 11 01472 2013.
ISSN: ISSN 1544-9173
PubMed: 23382649
DOI: 10.1371/JOURNAL.PBIO.1001472
Page generated: Tue Aug 19 08:08:04 2025
ISSN: ISSN 1544-9173
PubMed: 23382649
DOI: 10.1371/JOURNAL.PBIO.1001472
Last articles
W in 1DV4W in 1FR3
W in 1GUG
W in 1H9R
W in 1H9K
W in 1H0H
W in 1FEZ
W in 1FKA
W in 1E3P
W in 1E18